※ User Guide:
1. Q: How to use GPS-Uber webserver?
A: First, you can find the prediction website in 'WEB SERVER' page of GPS-Uber (http://gpsuber.biocuckoo.cn/online.php). Second, follow the procession in context of 'How to use GPS-Uber for protein/peptide ubiquitination prediction'. Wait a moment, you can get the results of general or E3-specific substrate ubiquitination sites.
2 . Q: How to read the GPS-Uber results?
A: Here we use the human protein RAC1 of RING/F-box E3 families as the example. After clicking "Submit", the prediction results of RING-catalyzed and F-box-catalyzed ubiquitination sites with prediction score are shown as follows:
<1>. The table of the GPS-Uber results (Page 1)
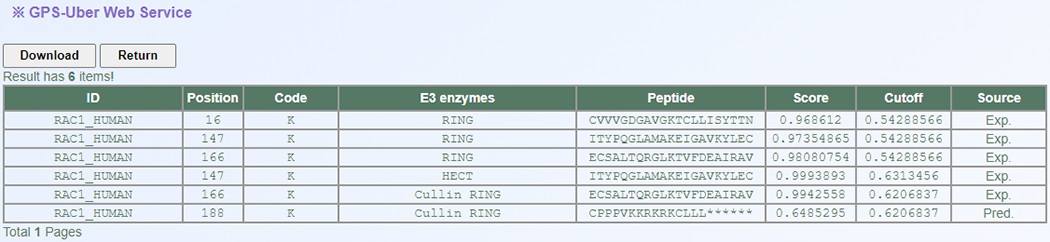
ID: The name/id of the protein sequence that you input to predict.
Position: The position of the site which is predicted to be ubiquitinated.
Code: The residue which is predicted to be ubiquitinated.
E3 enzymes: The regulatory E3 enzymes which is predicted to ubiquitinate the site.
Peptide: The predicted ubiquitinated peptide with 10 amino acids upstream and 10 amino acids downstream around the modified residue.
Score: The value calculated by E3-specific model to evaluate the potential of ubiquitination. The higher the value, the more potential the residue is ubiquitinated.
Cutoff: The cutoff value under the threshold. Different threshold means different precision, sensitivity and specificity.
Source: Whether this ubiquitination site validated by experiment, "Exp." means YES, while "Pred." means NO.
3 . Q: How to get the datasets used in GPS-Uber?
A: Here we provide the dataset for general prediction : Benchmark Data, Secondary Training Data, Testing Data.
4 . Q: I have a few questions which are not listed above, how can I contact the authors of GPS-Uber?
A: Please contact the authors through email for details. Thanks!

